Entry information : OsPrx46(LOC_Os03g25340)
| Entry ID | 1056 |
|---|---|
| Creation | 2005-07-20 (Filippo Passardi) |
| Last sequence changes | 2005-07-20 (Filippo Passardi) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2010-08-03 (Marie Brette (Scipio)) |
Peroxidase information: OsPrx46(LOC_Os03g25340)
| Name | OsPrx46(LOC_Os03g25340) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx143] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Viridiplantae (green plants); Streptophyta; Angiospermae; Monocotyledons | ||||||||||||||||||||||||||||||||||||
| Organism | Oryza sativa ssp japonica cv Nipponbare [TaxId: 39947 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | Roots |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
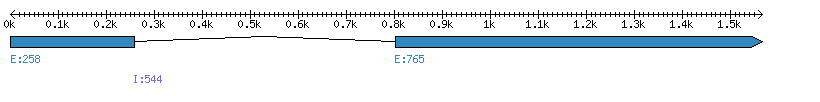
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references OsPrx46(LOC_Os03g25340)
| Literature | Passardi F., Longet, D., Penel C. and Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants Phytochemistry, 65 (13), 1879-1893 (2004) |
|---|---|
| Protein ref. | UniProtKB: Q5U1P7 |
| DNA ref. | GenBank: NC_008396.1 (14313840..14315403) |
Protein sequence: OsPrx46(LOC_Os03g25340)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | chromosome 3. 1 EST (young root), sequence of the EST is different from the one given in this database between AAMLRT and SGNDPT. The sequence in this database (predicted from BAC AC082644) is probably the correct one, as the one given by the EST generates a peroxidase without a critical CP in this precise region. More ESTs are needed to definitely conclude for a sequencing error. Note that there is no splicing at this site (middle of an exon!). | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
