Annotation Procedures
Three "annotation procedures" have been set up to produce expert and exhaustive data which will be included into the
RedoxiBase.
Each one is based on existing databanks or libraries (i) and on it's corresponding screening methods (ii).
It will produce different types of data (iii) which will be controlled manually before being integrated into the database (iv).
This last step is what sets apart the procedures done on the RedoxiBase from other annotation procedures.
The fact that the resulting sequences are revised thoroughly increases greatly the quality of the sequences present in this database.
Below is a schematic description of the annotation procedures done in order to add new sequences into the RedoxiBase:
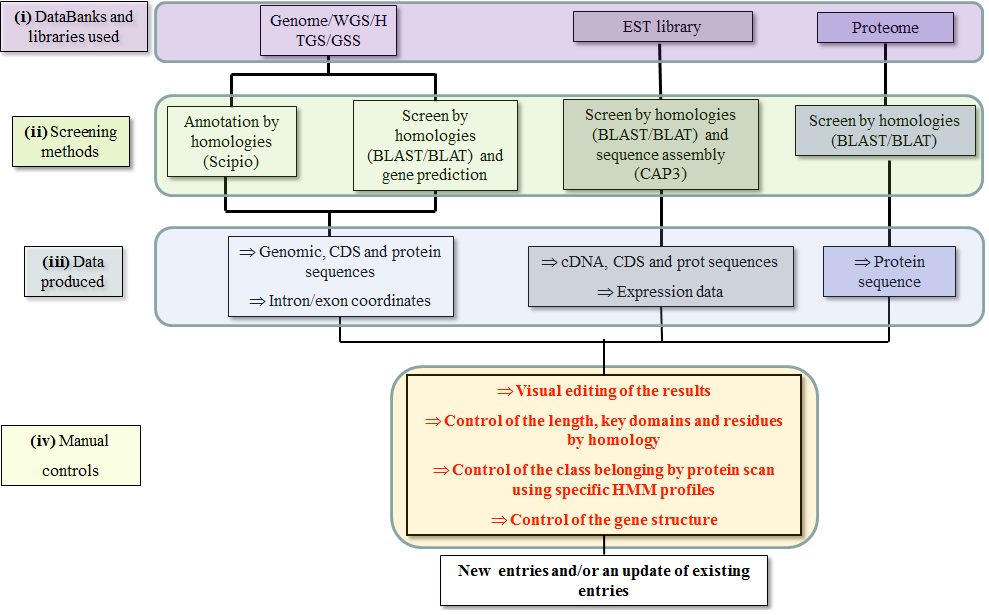
Fig. Annotation procedures.
Each one is based on existing databanks or libraries (i) and on it's corresponding screening methods (ii).
It will produce different types of data (iii) which will be controlled manually before being integrated into the database (iv).
This last step is what sets apart the procedures done on the RedoxiBase from other annotation procedures.
The fact that the resulting sequences are revised thoroughly increases greatly the quality of the sequences present in this database.
Below is a schematic description of the annotation procedures done in order to add new sequences into the RedoxiBase:

Fig. Annotation procedures.
Nomenclature of the input sequence
Organism abbreviation (Org) and Class abbreviation (Class) => the name entry will be OrgClass#
- For alternative splicing the name entry will be: OrgClass#-A, B, C
- For gene duplication the name entry will be: OrgClass#-1, -2, -3
- In the particular situation of polyploid genome with known origine of the different chromosome, the name entry of duplicated genes can be OrgClass#-1A, OrgClass#-1B
