Entry information : MtPrx01 (Rip1 / Medtr5g074860.1[4.0] / Medtr5g074860.1[3.5] / Medtr5g082780.1[3.0])
| Entry ID | 59 |
|---|---|
| Creation | 2006-09-06 (Christophe Dunand) |
| Last sequence changes | 2010-09-14 (Haythem Mhadhbi) |
| Sequence status | complete |
| Reviewer | Bruno Savelli |
| Last annotation changes | 2017-08-08 (Bruno Savelli) |
Peroxidase information: MtPrx01 (Rip1 / Medtr5g074860.1[4.0] / Medtr5g074860.1[3.5] / Medtr5g082780.1[3.0])
| Name (synonym) | MtPrx01 (Rip1 / Medtr5g074860.1[4.0] / Medtr5g074860.1[3.5] / Medtr5g082780.1[3.0]) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx010] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Fabaceae Medicago | ||||||||||||||||||||||||||||||||||||
| Organism | Medicago truncatula (barrel medic) [TaxId: 3880 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue types | Immature seeds Pods Roots |
||||||||||||||||||||||||||||||||||||
| Inducers | Pathogen interaction Phytophthora infection Rhizobium inoculation |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
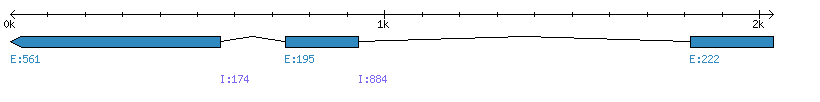
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references MtPrx01 (Rip1 / Medtr5g074860.1[4.0] / Medtr5g074860.1[3.5] / Medtr5g082780.1[3.0])
| Literature | Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7 (1), 43-55 (1995) |
|---|---|
| Protein ref. | UniProtKB: Q40372 |
| DNA ref. | Phytozome 12: chr5 (31799523..31797488) |
| Cluster/Prediction ref. | Phytozome Gene 12: 31090495 UniGene: Mtr.12647 |
| Omic ref. | Gene atlas: Mtr.25211.1.S1_at Legoo: Mtr.25211.1.S1_at |
| Other ref. | Medicago_jcvi: 23235 |
Protein sequence: MtPrx01 (Rip1 / Medtr5g074860.1[4.0] / Medtr5g074860.1[3.5] / Medtr5g082780.1[3.0])
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 5, introns 1 and 2), 26 ESTs. Transcript variant predicted by Phytozome (extra intron and lack of VALS). | ||||||||||||
| Promoter► Send to BLAST Send to cis Analysis |
|
||||||||||||
| Terminator +► Send to BLAST Send to cis Analysis |
|
||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
| cDNA► Send to BLAST |
|
||||||||||||
