Entry information : SmPrxQ-1_21 (SmPrxQa_21)
| Entry ID | 6219 |
|---|---|
| Creation | 2008-07-07 (Christophe Dunand) |
| Last sequence changes | 2012-04-26 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2012-04-26 (Christophe Dunand) |
Peroxidase information: SmPrxQ-1_21 (SmPrxQa_21)
| Name (synonym) | SmPrxQ-1_21 (SmPrxQa_21) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Atypical 2-Cysteine peroxiredoxin (type Q) [Orthogroup: PrxQ001] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Isoetopsida Selaginellaceae Selaginella | ||||||||||||||||||||||||||||||||||||
| Organism | Selaginella moellendorffii [TaxId: 88036 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
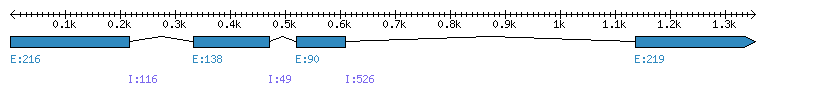
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references SmPrxQ-1_21 (SmPrxQa_21)
| Literature | Richardson,P., Lucas,S., Rokhsar,D., Wang,M. and Lindquist,E.A. DOE Joint Genome Institute Selaginella moellendorffii EST project. Unpublished (2008). |
|---|---|
| DNA ref. | Phytozome 12: scaffold_21 (1420112..1421465) |
| mRNA ref. | Phytozome 12: 148953 [Incorrect prediction] |
| EST ref. | GenBank: FE446757.1 |
| Cluster/Prediction ref. | UniGene: Smo.828 |
Protein sequence: SmPrxQ-1_21 (SmPrxQa_21)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (3 introns) and 32 ESTs. Unigene compiles ESTs from both haplotypes. incorrect prediction from Phytozome (first exon is missing) | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
| cDNA► Send to BLAST |
|
||||||||||||
