Entry information : CpurNOx01 (CpNox1)
| Entry ID | 7619 |
|---|---|
| Creation | 2010-09-29 (Christophe Dunand) |
| Last sequence changes | 2010-09-29 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2010-09-30 (Christophe Dunand) |
Peroxidase information: CpurNOx01 (CpNox1)
| Name (synonym) | CpurNOx01 (CpNox1) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Ancestral NADPH oxidase [Orthogroup: NOx002] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Ascomycota Sordariomycetes Clavicipitaceae Claviceps | ||||||||||||||||||||||||||||||||||||
| Organism | Claviceps purpurea (ergot fungus) [TaxId: 5111 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
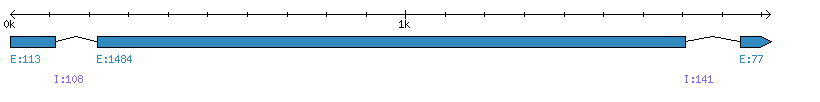
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references CpurNOx01 (CpNox1)
| Literature | Giesbert,S., Schuerg,T., Scheele,S. and Tudzynski,P. The NADPH oxidase Cpnox1 is required for full pathogenicity of the ergot fungus Claviceps purpurea. MOLECULAR PLANT PATHOLOGY (2008), 9(3), 317–327. |
|---|---|
| DNA ref. | GenBank: AM899998.1 (366..2288) |
Protein sequence: CpurNOx01 (CpNox1)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (2 introns). | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
