Entry information : AmusPrxV
| Entry ID | 11672 |
|---|---|
| Creation | 2013-02-28 (Christophe Dunand) |
| Last sequence changes | 2013-02-28 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2013-07-26 (Catherine Mathe (Scipio)) |
Peroxidase information: AmusPrxV
| Name | AmusPrxV | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Atypical 2-Cysteine peroxiredoxin (type V) [Orthogroup: PrxV001] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Basidiomycota Agaricomycetes Amanitaceae Amanita | ||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Amanita muscaria [TaxId: 41956 ] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||
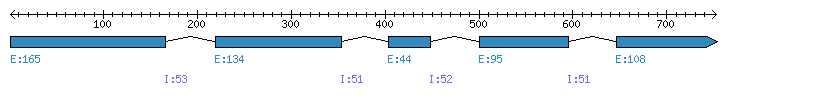
| Gene structure Fichier |
Exons►
|
||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references AmusPrxV
| DNA ref. | JGI genome: scaffold_112 (68929..69681) |
|---|---|
| Cluster/Prediction ref. | JGI gene: 1024964 |
Protein sequence: AmusPrxV
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (4 introns). | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
