Entry information : PcCIIBA01 (PcNOP01 / nop-a / nop-b)
| Entry ID | 2332 |
|---|---|
| Creation | 2007-02-07 (Christophe Dunand) |
| Last sequence changes | 2011-11-24 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2015-04-29 (Christophe Dunand) |
Peroxidase information: PcCIIBA01 (PcNOP01 / nop-a / nop-b)
| Name (synonym) | PcCIIBA01 (PcNOP01 / nop-a / nop-b) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Other class II peroxidase type A [Orthogroup: CIIBA001] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Basidiomycota Agaricomycetes Corticiaceae Phanerochaete | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Phanerochaete chrysosporium [TaxId: 5306 ] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
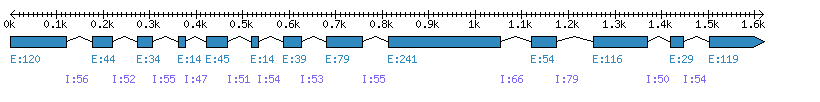
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references PcCIIBA01 (PcNOP01 / nop-a / nop-b)
| Literature | REFERENCE 1 Larrondo,L.F., Gonzalez,A., Perez-Acle,T., Cullen,D. and Vicuna,R. The nop gene from Phanerochaete chrysosporium encodes a peroxidase with novel structural features. Biophys Chem 2005 Jul 1, 116(2): 167-173.
REFERENCE 2 Martinez,D., Larrondo,L.F., Putnam,N., Sollewijn Gelpke,M.D., Huang,K., Chapman,J., Helfenbein,K.G., Ramaiya,P., Detter,J.C., Larimer,F., Coutinho,P.M., Henrissat,B., Berka,R., Cullen,D. and Rokhsar,D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22 (6), 695-700 (2004). |
|---|---|
| Protein ref. | UniProtKB: Q5XXE3 Q5XXE5 [Mismatch] Q5XXE2 [Other strain] |
| DNA ref. | JGI genome: scaffold_10 (1250082..1251701) |
| mRNA ref. | GenBank: AY727765 |
Protein sequence: PcCIIBA01 (PcNOP01 / nop-a / nop-b)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic and mRNA. Strain="BKM1767", "RP-78" and "ME446". No EST. The 3'end looks shorter with missing conserved residues. This class II peroxidase has been called "NoP", i.e. "novel peroxidase", by its discoverers. It is considerably shorter than other class II peroxidases. Two SwissProt Q5XXE3 (nop-b) and Q5XXE5 (nop-a) submitted by the same authors (differs by a few amino acids). There is only ONE nop gene in the P.chrysosporium genome (BLAST in Joint Genome Institute). |
||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
